Improving Parallel Performance
We all want better performance, don't we? Since parallel molecular dynamics involves large amounts of communication between nodes for each step, the underlying hardware and MPI library will have a very direct effect on communication and thus on your total simulation performance. Small systems (less than 10,000 atoms) are usually most sensitive to the latency, while larger simulations (100,000 atoms or more) also need high bandwidth to send all the data each step. This section is a summary of some of the things we've learned from our own cluster experience, many of them are not specific to Gromacs, but should apply to parallel programs in general. You can safely skip it if you are a novice, but read on if you are building a new cluster or would like your current network to perform better.Hardware Considerations
Gromacs comes with manually tuned assembly loops for a wide range of hardware. AMD Opteron CPUs currently provide the best price/performance ratio, closely followed by IBM PowerPC 970 (Apple G5) and Intel Xeon/Pentium IV. If you are building your own cluster for the first time it might still be a good idea to use Xeon, simply because it is the oldest and most tested architecture. We strongly recommend that you use SMP (dual) machines in the cluster since the shared-memory communication between the CPUs has extremely low latency and high bandwidth -- and they are cheaper than two single-CPU boxes.For SMP Xeons, it is important to turn OFF hyperthreading in the bios. This might seem like incredibly stupid advice, since four processors should be better than two, right? Unfortunately, the Linux kernel (including version 2.6) still has problems to do proper scheduling with hyperthreading: If you run two jobs, they sometimes end up on the two virtual CPUs sharing inside one physical CPU, while the other physical CPU is idle. You can imagine what this does to your performance, so just say no to hyperthreading for now. Even if the Linux scheduler did a perfect job you would not see much difference for CPU-bound programs like Gromacs - the integer and floating-point units are not duplicated in current Xeon implementations.
It is a good idea to have some sort of network between the nodes in your cluster. If you have a deep wallet or can apply for time at existing supercomputer facilities, the best solution is to use a dedicated high-performance interconnect such as Myrinet, Infiniband or Scali that provide both low latency and high bandwidth. All these vendors provide their own MPI libraries and documentation, so in that case you should stop reading this and instead ask the vendor how to get the best possible performance. If you are spending your own funds the situation is a little more complex. Myrinet or the alternatives might still be worth it in some cases, but due to the fairly high price we often accept standard Gigabit Ethernet so we can buy more nodes instead.
Tweaking Your MPI Library
What if you already have a cheap Ethernet-based cluster that you would like to be faster, but you're not willing to spend a single cent? Fortunately there are some free lunches in the Free Software world! MPI over Ethernet is slow because it usually involves copying data to a temporary buffer and then calling the Linux network card drivers, and vice versa at the receiving node. If the message is larger than the "small message" buffer size we must first send a header message warning that a large message will follow. This is portable and resource-efficient, but you can probably imagine what it does to your performance. Let's see what can be done about it. First, if you are running Linux there are special MPI implementations (MPI/GAMMA or Scali MPI Connect) that can talk directly to a small number of network cards and bypass the driver. Check them out with Google and compare to your hardware. If your system is supported you could achieve latencies as low as 10 microseconds!Unfortunately the special libraries do not support the gigabit cards in our nodes, so we have to stick with LAM/MPI, which we have found to be a tiny bit faster than MPICH. You may want to explore the next generation MPI's such as MPICH2 and Open MPI as well. However, even in the case of LAM/MPI it is well worth recompiling the library with better options. By default TCP/IP communication is used between all processes, but you most certainly want to use shared memory instead when two processes are on the same node. You should also increase the âsmall messageâ buffer size significantly. Download the source from the LAM site and configure it as
./configure -prefix=/home/joe/software \
-with-rpi=usysv \
-with-tcp-short=524288 \
-with-shm-short=524288
Here we assumed you do not have root privileges, but want to install the software under your home directory /home/joe. Issue make and then make install, and your new tweaked MPI library is ready to use once you add /home/joe/software/bin to your path.
Your Batch Queue System
Ideally the batch system should optimize the allocation if you for instance ask for 8 processors, but in practice it is more or less random. You want to make sure the 8 processors are confined 4 SMP nodes where you have both CPUs, and in a large cluster with one switch per rack all the nodes should be in the same rack so you only have a single switch between them. For the PBS and Torque queue systems this can be accomplished with a batch submission command likeqsub -l nodes=4:rack1:ppn=2 script
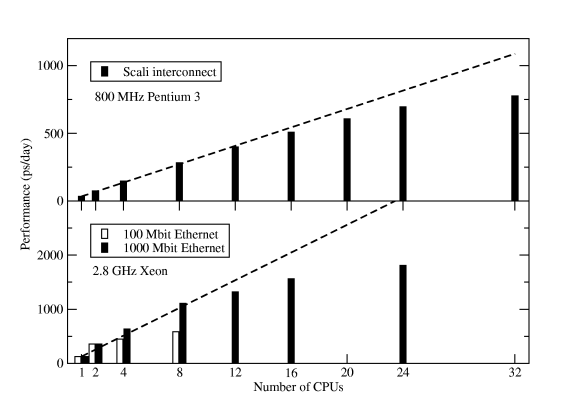
This command only works if the property ârack1â is assigned to all nodes in rack1. If it's not done on your cluster, bug the administrator until it is fixed -- this is a big performance gain compared to running on single processors per node spread over two or more racks. Figure Six shows some results for the standard Gromacs scaling benchmark -- a 130,000-atom DPPC membrane system with long electrostatics cut-offs.
Conclusions
The simulation process described here is quite typical: an initial energy minimization followed by equilibration with position restraints, although the equilibration could be a bit longer in practice. Since the first part of the non-restrained simulation should also be discarded as equilibration in practice, it is common to perform another free simulation with pressure coupling (to achieve the right density) before the production run starts, but we simply didn't have room for that in this tutorial!When you start your own simulations, remember to be gentle with your proteins and don't neglect warning messages -- there is a reason why they are there. Unfortunately simulations are actually very much like laboratory experiments: it is very easy to destroy months of work with a moment of recklessness. There is a wealth of information available at the Gromacs website, including a paper manual and more important: several very active user mailing list. We're looking forward to see your posts there!
This article was originally published in ClusterWorld Magazine. It has been updated and formatted for the web. If you want to read more about HPC clusters and Linux you may wish to visit Linux Magazine.
Erik Lindahl is a Gromacs co-author and assistant professor at the Stockholm Bioinformatics Institute, Sweden.


